NSPA: characterizing the disease association of multiple genetic interactions at single-subject resolution
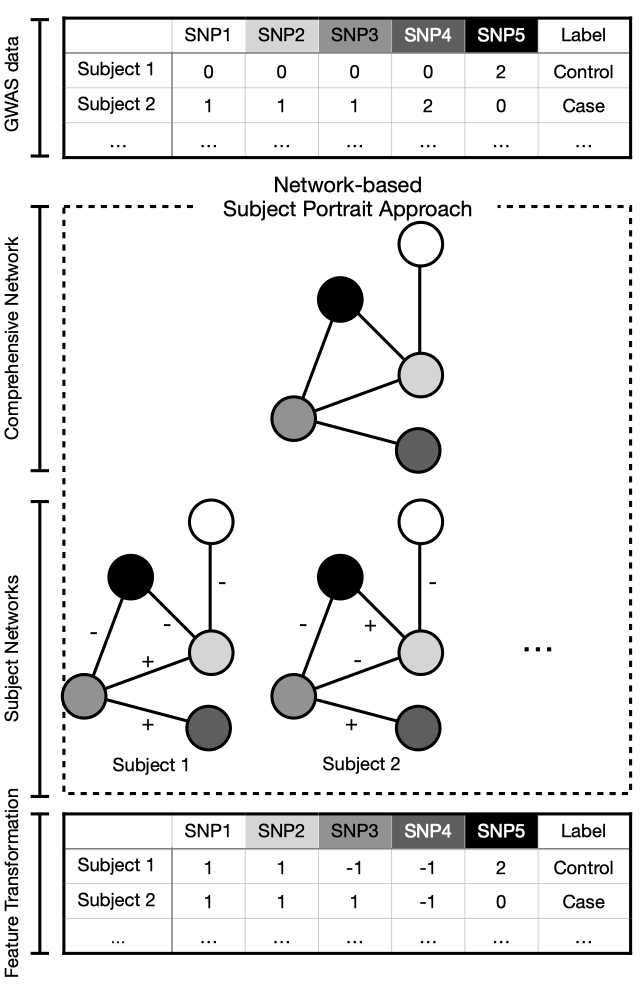
We propose the Networkbased Subject Portrait Approach (NSPA) and an accompanying feature transformation method to determine the collective risk impact of multiple genetic interactions for each subject. [Link]
Authors:
- Zhendong Sha
- Dr. Yuanzhu Chen
- Dr. Ting Hu
Abstract
Motivation: The interaction between genetic variables is one of the major barriers to characterizing the genetic architecture of complex traits. To consider epistasis, network science approaches are increasingly being used in research to elucidate the genetic architecture of complex diseases. Network science approaches associate genetic variables’ disease susceptibility to their topological importance in the network. However, this network only represents genetic interactions and does not describe how these interactions attribute to disease association at the subject-scale. We propose the Networkbased Subject Portrait Approach (NSPA) and an accompanying feature transformation method to determine the collective risk impact of multiple genetic interactions for each subject.
Results: The feature transformation method converts genetic variants of subjects into new values that capture how genetic variables interact with others to attribute to a subject’s disease association. We apply this approach to synthetic and genetic datasets and learn that 1) the disease association can be captured using multiple disjoint sets of genetic interactions; 2) the feature transformation method based on NSPA improves predictive performance comparing with using the original genetic variables. Our findings confirm the role of genetic interaction in complex disease and provide a novel approach for gene-disease association studies to identify genetic architecture in the context of epistasis.
Availability and implementation: The codes of Network-based Subject Portrait Approach are now available in: https://github.com/MIB-Lab/Network-based-Subject-Portrait-Approach.
Keywords
Genetic networks, Genetics, Genome analysis, Network analysis
How it works
This work was accepted by Bioinformatics Advances